courses
I offer various courses distilling the most important aspects of software engineering for scientific groups. Courses can be in-person or online, and they usually consist of a presentation part and a workshop part, but can be fully customized to your needs. I have so far given courses at:
- Technical University of Munich
- University of California, Berkeley
Do not hesitate to reach out and we can discuss potential courses for your group! Just write me an e-mail.
Topics
The topics of the courses can be adjusted to your particular needs. From my experience, the Best practices for coding in science and Fundamentals of data science courses are highly useful for most research groups. Even though it is mostly a presentation, is it still interactive, with quizzes and small tasks. Notably, I have recently also started showing how to use AI tools effectively in technical tasks. In my opinion, universities are far behind what could be done here.
The other courses contain both presentation and hands-on workshop parts.
But generally, we can adapt the courses as you need, as potentially not all aspects will be relevant for your group. Here are some general courses that I have taught:
Courses
- Best practices for programming for scientists
- Beyond vibe-coding: programming successfully with AI
- Version control with
gitfor scientists - Monitoring and optimizing resource usage of scientific code
- Making quantitative research reproducible
- Fundamentals of data science
- Fundamentals of programming in Python
More details on the courses
Best practices for programming for scientists (2 x 2h presentation)
- Coding pitfalls (classic bugs you need to know about)
- Programming paradigms (writing maintainable code)
- Using Integrated Development Environments (IDEs) for fast and efficient programming
- Debugging code
- Version control with
git - Testing your code
- Leveraging AI tools
See also my blog posts on writing proper code and programming paradigms.
Beyond “vibe-coding”: programming successfully with AI (~3h presentation, potentially with workshop)

AI is here to stay and you’d have a competitive disadvantage if you didn’t use it. But anyone can ask ChatGPT to write them some code. But do you just “vibe-code” or use the tools at hand efficiently? In this workshop, we will look into:
- How to efficiently program with AI tools
- How to make sure AI-generated or AI-influenced code is correct?
- How can teachers detect AI-generated code?
Version control with git for scientists (integrated presentation and workshop, 4h total)
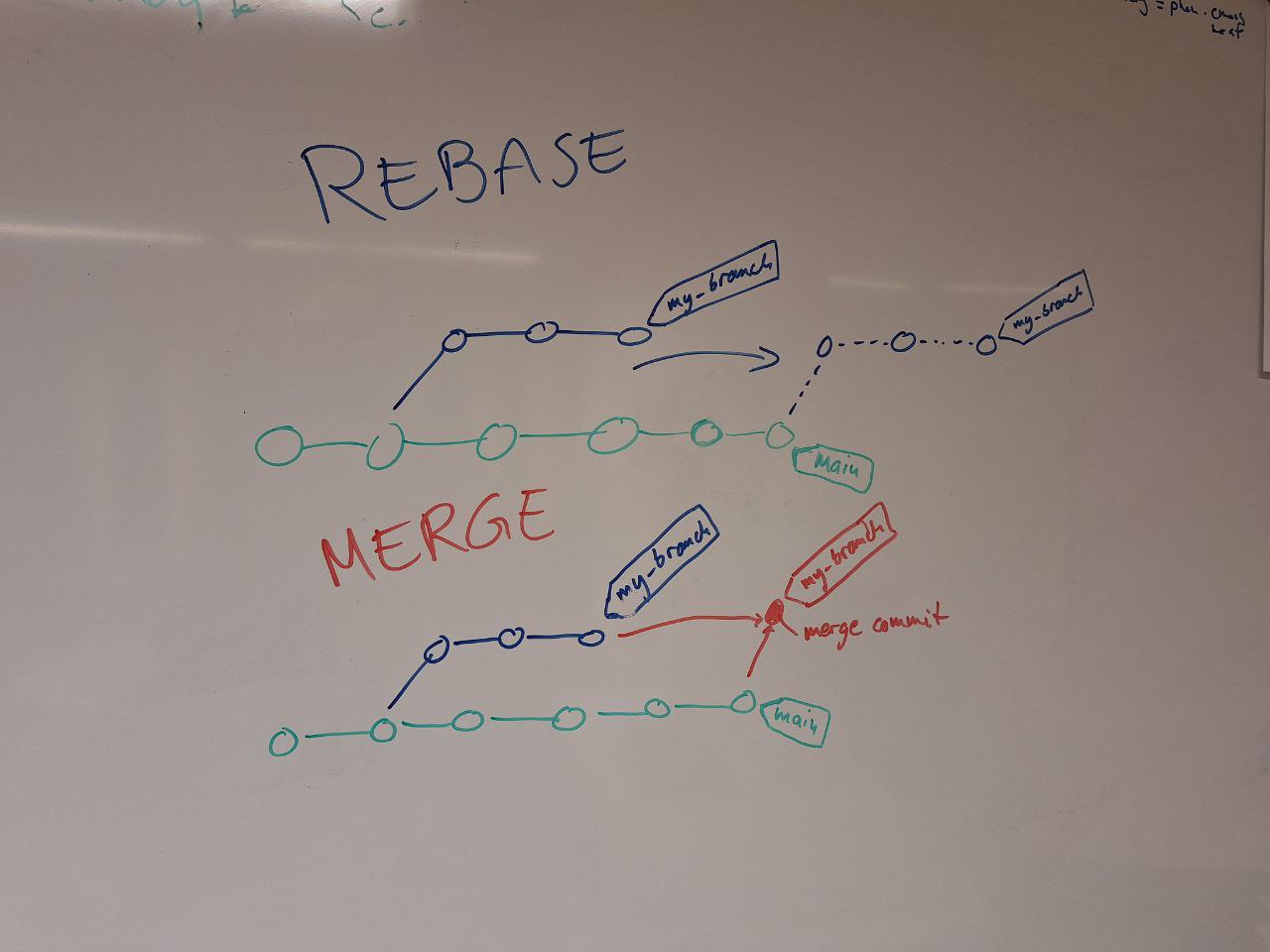
- What are the benefits of version control and why should all code be in version control?
- What is
git, what isGithub? - Setting everything up
- Understanding the benefits of version control and how to make use of it
- How to properly use
git- commits
- branches
- merging
- checking what has changed
You can find the content of the workshop here: https://github.com/k-gregor/git-workshop
Monitoring and optimizing resource usage of scientific code (~3h presentation, potentially with workshop)
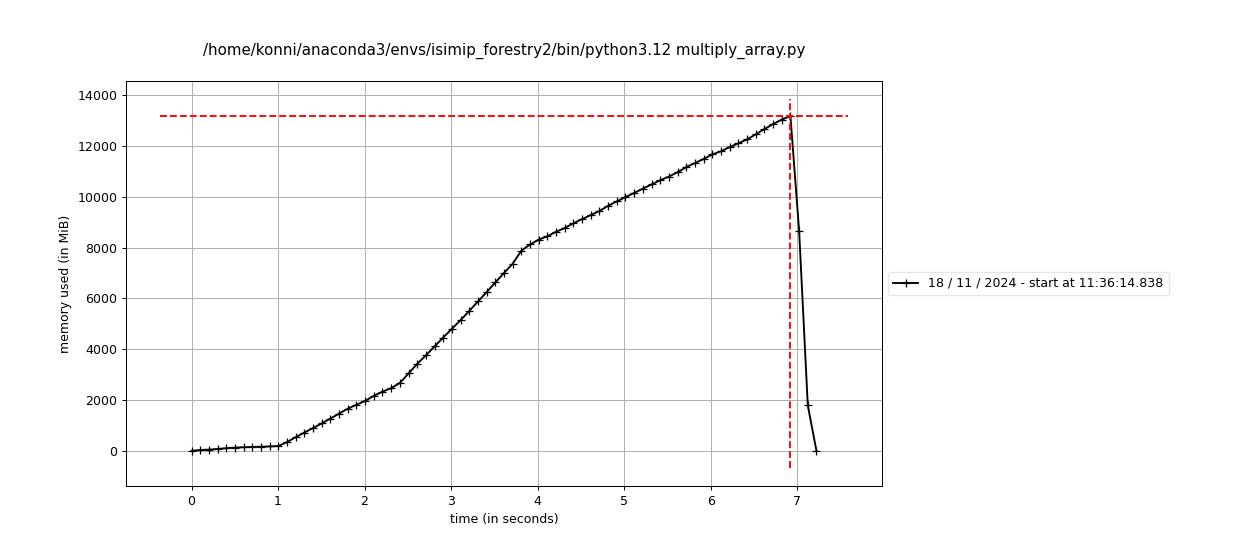
- Understanding the memory architecture of computers
- How to monitor total usage of computer programs
- Professional profiling tools
- A brief introduction to data structures and runtime analysis (O-notation)
- Programming memory-efficiently
- chunking
- data types
- lazy loading
- in-place operations
See also my blog post on this topic: memory aspects in scientific code
Using the command line and bash scripting to speed up scientific workflows (integrated presentation and workshop, 4h total)
This covers topics that are helpful both when working locally or on a supercomputer.
- Navigating through files and directories
- Finding data quickly
- Superfast data manipulation in the command line
- Writing scripts to automate workflows
- Monitoring resource usage of data analyses
Introduction to Continuous Integration and Continuous Deployment (~1h presentation)
- Collaborating
- Automated documentation
- Code linting
- Automated testing
- Issue tracking
- Versioning
Making quantitative research reproducible (Presentation 2h, potentially with workshop)
- Making scientific workflows reproducible with
snakemake(Python) ortargets(R) - Dockerize your code to make it run anywhere